Adding Custom Channels to DeepVariant
Authors:
Background
DeepVariant is a deep-learning based variant caller that formulates variant calling as an image classification problem. Aligned sequences are read from a BAM file, and DeepVariant extracts a series of sequence features from read pileups and converts them to a corresponding set of input images that we call channels. Each channel encodes a sequence feature extracted from reads. These channels are the fundamental unit by which we feed data into our model, and we combine a series of them as input to train our model to accurately predict genotypes.
All DeepVariant production models use these 6 core channels (Figure 1):
- read base
- base quality
- mapping quality
- strand of alignment
- read supports variant
- base differs from ref
These get concatenated into a single image and are used as input into DeepVariant models.

New sequencing technologies may produce novel signals (e.g. raw basecalling metrics) that could be encoded as a channel to improve accuracy specific to that platform. Alternatively, custom channels can specify derived properties (e.g. GC content) that can be experimented with to improve model accuracy on existing platforms.
To allow models to capture additional signals, we introduced a framework for developing custom channels in DeepVariant 1.2. Custom channels are specified as a list using the --channels flag with make_examples (see the original Nature Biotechnology paper for an introduction to how DeepVariant works). Each custom channel can encode novel signals or derived sequence properties that might help DeepVariant make more accurate predictions. This blog post will cover how to develop a new custom channel, how to train a model for that channel, and how to run inference using a model that incorporates a custom channel.
We have successfully used the new framework with the release of DeepVariant v1.4 to include a new custom channel called insert_size which encodes the fragment length of Illumina paired end sequencing reads. Our analysis finds that inclusion of the insert_size channel in our models results in models that are 4.7% more accurate on WGS and 9.8% on WES.
In another recent development, Ultima Genomics has put together custom channels for their new sequencing platform. The Ultima Genomics sequencing platform produces probabilities for error in homopolymer length (encoded in the base-quality string, together with an auxiliary field called TP) and a probability score for missed signals (T0). These signals are incorporated as custom channels and lead to improved variant calling accuracy with their platform.
This tutorial will show you how to add custom channels to DeepVariant, train new models, and use them to perform variant calling. Users are expected to have intermediate experience with C++ and Python.
How are Channels Constructed?
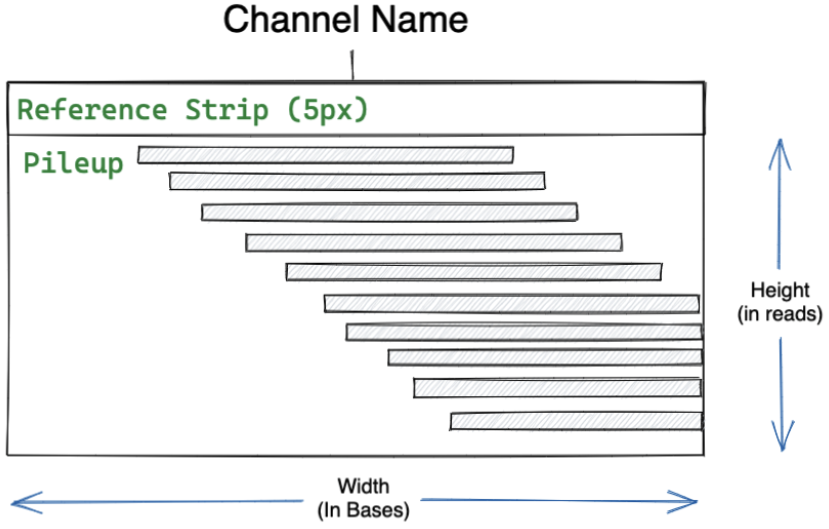
Although we visualize channels as images, their underlying representation is as multidimensional tensor objects. Each channel is encoded as a matrix of values ranging from 0-255. Channels have two components (Figure 2). The top five pixels are known as the “reference strip”, and can be used to specify information derived from the reference, although only a few channels use this component (e.g. read base). Below the reference strip is the read pileup. Given that channels are encoded using pileup information, the width corresponds to a set number of bases whereas the height reflects the maximum number of reads. While these parameters are configurable, experimentation has yielded the optimal width and height for short paired end sequence reads to be 221 and 100, respectively. make_examples generates these matrices, and stacks them together to form 3D tensor objects as model input.
To build a custom channel, we’ll have to define some functions and logic to extract the desired sequence features using the Nucleus library, and scale their values appropriately within a 0-255 value range. There are two channel types currently:
- read-level channels - assign a single value to an entire read within the pileup (e.g. read supports variant).
- base-level channels - base-level channels specify values at the base level (e.g. base quality).
How to Add a New Custom Channel
The process of adding an Optchannel will require forking DeepVariant, allowing you to make edits to the codebase. The following example will add a new channel called read_orientation that encodes information about the orientation of paired end read alignments. Once the channel has been added, we will describe how to train a model and apply it to call variants using this new channel.
git clone https://github.com/google/deepvariant.git
cd deepvariant
1. Declare a new channel option
deepvariant/dv_constants.py
The OPT_CHANNELS list defines the set of available custom channels that can be used by make_examples.py, so we will begin by naming our channel and adding it to this list.
# Define available OptChannels (optional extra channels).
OPT_CHANNELS = [
'read_mapping_percent', 'avg_base_quality', 'identity',
'gap_compressed_identity', 'gc_content', 'is_homopolymer',
'homopolymer_weighted', 'blank', 'insert_size','read_orientation',
]
2. Add this channel name to the DeepVariantChannelEnum
deepvariant/protos/deepvariant.proto
This proto is used to help the python and C++ components of DeepVariant communicate with one another. By convention, enum values are prefixed with CH_ and uppercased. We’ll add an enum value to represent the new read orientation channel. Custom user channels should use enum values above 1000 so that it does not collide with future channel additions by the DeepVariant team.
enum DeepVariantChannelEnum {
// Default should be unspecified.
CH_UNSPECIFIED = 0;
// 6 channels that exist in all DeepVariant production models.
CH_READ_BASE = 1;
CH_BASE_QUALITY = 2;
CH_MAPPING_QUALITY = 3;
CH_STRAND = 4;
CH_READ_SUPPORTS_VARIANT = 5;
CH_BASE_DIFFERS_FROM_REF = 6;
…
// The following channels correspond to the "Opt Channels" defined in
// deepvariant/pileup_channel_lib.h:
CH_READ_MAPPING_PERCENT = 11;
CH_AVG_BASE_QUALITY = 12;
CH_IDENTITY = 13;
CH_GAP_COMPRESSED_IDENTITY = 14;
CH_GC_CONTENT = 15;
CH_IS_HOMEOPOLYMER = 16;
CH_HOMEOPOLYMER_WEIGHTED = 17;
CH_BLANK = 18;
CH_INSERT_SIZE = 19;
CH_READ_ORIENTATION = 1000;
}
3. Add the channel name as a constant in our OptChannel library file
deepvariant/pileup_channel_lib.h
This pileup_channel_lib.h file contains the logic for drawing custom channels. We will first define a value for the read_orientation channel to help bridge the gap between Python and C++.
//--------------//
// Opt Channels //
//--------------//
static const auto& ch_read_mapping_percent = "read_mapping_percent";
static const auto& ch_avg_base_quality = "avg_base_quality";
static const auto& ch_identity = "identity";
static const auto& ch_gap_compressed_identity = "gap_compressed_identity";
static const auto& ch_gc_content = "gc_content";
static const auto& ch_is_homopolymer = "is_homopolymer";
static const auto& ch_homopolymer_weighted = "homopolymer_weighted";
static const auto& ch_blank = "blank";
static const auto& ch_insert_size = "insert_size";
static const auto& ch_read_orientation = "read_orientation";
4. Map this channel name to the DeepVariantChannelProto enum
deepvariant/pileup_channel_lib.h
A simple function is used to map the channel name to its corresponding enum. So we will follow the pattern of existing channels in this function for the read_orientation channel.
inline DeepVariantChannelEnum ChannelStrToEnum(const std::string& channel) {
// Maps channel string representation to DeepVariantChannelEnum
if (channel == ch_read_mapping_percent)
return DeepVariantChannelEnum::CH_READ_MAPPING_PERCENT;
if (channel == ch_avg_base_quality)
return DeepVariantChannelEnum::CH_AVG_BASE_QUALITY;
...
if (channel == ch_read_orientation)
return DeepVariantChannelEnum::CH_READ_ORIENTATION;
CHECK(false) << "Channel '" << channel << "' should have a corresponding "
<< "enum in DeepVariantChannelEnum.";
}
5. Define the channel encoding
deepvariant/pileup_channel_lib.h
This is naturally the most important step as it specifies how to encode your channel. Whatever data you are interested in representing, here is where you map it to a pixel value from 0 to 255. In our case, we will take 4 bitwise flags present in the FLAG section of a given read in a BAM file, and map them to a single pixel value per read.
// Encodes read orientation and alignment information of read
inline std::vector<uint8> ReadOrientation(const Read& read) {
/* Maps the bit fields `proper_placement`, `secondary_alignment`,
* `supplementary_alignment` and `reverse_strand` into a 4-bit
* integer value, i.e 0-15:
*
* 0000000
* 0001000 <- proper_placement (8)
* 0010000 <- secondary_alignment (16)
* 0100000 <- supplementary_alignment (32)
* 1000000 <- alignment.position.reverse_strand (64)
*
*/
int value = 0;
if (read.proper_placement()) {
value += 8;
}
if (read.secondary_alignment()) {
value += 16;
}
if (read.supplementary_alignment()) {
value += 32;
}
if (read.alignment().position().reverse_strand()) {
value += 64;
}
std::vector<uint8> read_orientation_vector(read.aligned_sequence().size(),
value);
return read_orientation_vector;
}
6. Tell OptChannels.CalculateChannels to use your new Channel
deepvariant/pileup_channel_lib.h
class OptChannels {
public:
std::map<std::string, std::vector<unsigned char>> data_;
std::map<std::string, std::vector<unsigned char>> ref_data_;
void CalculateChannels(const std::vector<std::string>& channels,
const Read& read) {
// Calculates values for each channel
for (const std::string& channel : channels) {
if (channel == ch_read_mapping_percent) {
data_[channel].assign(
{ScaleColor(ReadMappingPercent(read), MaxMappingPercent)});
// ...
} else if (channel == ch_insert_size) {
data_[channel] = ReadInsertSize(read);
}
else if (channel == ch_read_orientation) {
data_[channel] = ReadOrientation(read);
}
}
}
…
}
7. Define how to represent the reference rows
deepvariant/pileup_channel_lib.h
Each pileup image has 5 reference rows at the top. You can define how these are represented in your channel in CalculateRefRows. For most custom channels we simply use 5 blank (255) rows.
void CalculateRefRows(const std::vector<std::string>& channels,
const std::string& ref_bases) {
// Calculates reference row values for each channel
// Create a fake read to represent reference bases.
Read refRead;
for (const std::string& channel : channels) {
if (channel == ch_read_mapping_percent) {
ref_data_[channel].assign({static_cast<uint8>(kMaxPixelValueAsFloat)});
// ...
} else if (channel == ch_read_orientation) {
ref_data_[channel].assign({static_cast<uint8>(kMaxPixelValueAsFloat)});
} else if (channel == ch_gc_content) {
// ...
}
}
}
That’s it! Now let’s see it in action.
Verifying a Channel was Added Correctly
Now that we have added code for generating our new channel, we can build a docker image to test it out.
DOCKER_IMAGE="deepvariant/custom-channel"
docker build -t ${DOCKER_IMAGE} .
It is a good idea to start by checking that DeepVariant can use our new channel during make_examples. Assuming we have prepared the same environment as the WES case study, we can declare the following variables:
CHANNEL="read_orientation"
REF=/reference/GRCh38_no_alt_analysis_set.fasta
BAM=/input/HG003.novaseq.wes_idt.100x.dedup.bam
BED=/input/idt_capture_novogene.grch38.bed
EXAMPLES=/output/HG002.chr20.${CHANNEL}.examples.tfrecord.gz
Then we may run make_examples.
docker run \
-v "${PWD}/input":"/input" \
-v "${PWD}/output":"/output" \
-v "${PWD}/reference":"/reference" \
${DOCKER_IMAGE} \
/opt/deepvariant/bin/make_examples \
--mode calling \
--ref ${REF} \
--reads ${BAM} \
--regions "chr20:0-500000" \
--examples ${EXAMPLES} \
--channels ${CHANNEL} \
--logtostderr
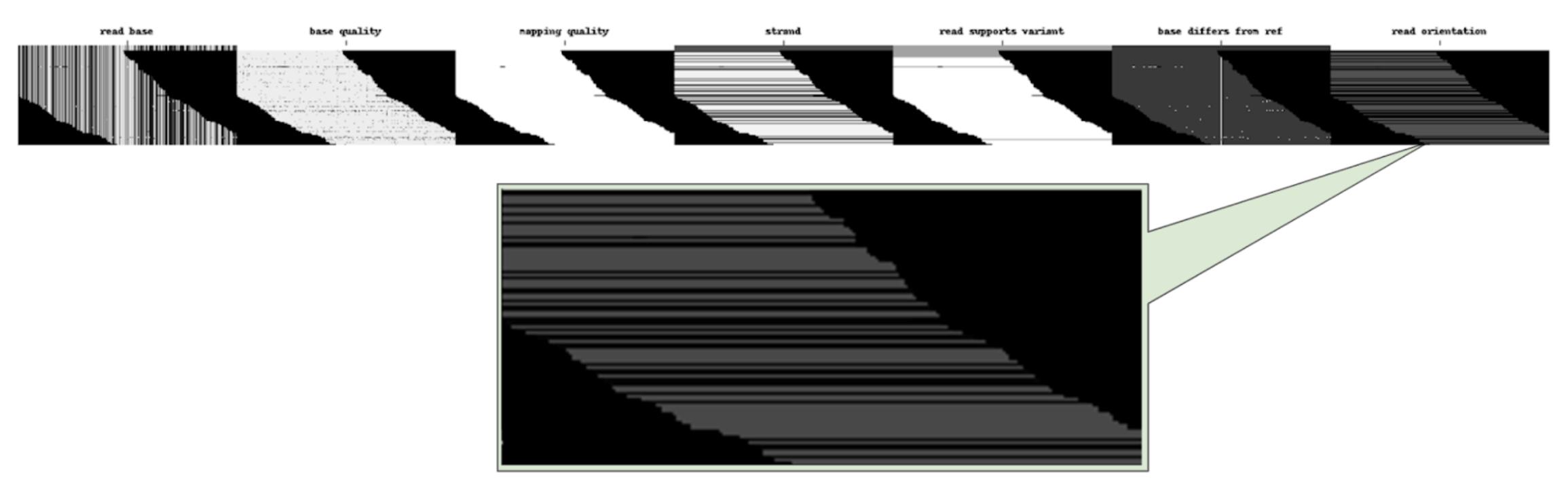
Assuming that DeepVariant found some candidates and created examples for them, we can use show_examples to visualize them and ensure our new channel is part of the pileup image.
docker run \
-v "${PWD}/output":"/output" \
${DOCKER_IMAGE_TAG} \
/opt/deepvariant/bin/show_examples \
--examples="${EXAMPLES}" \
--output="/output/" \
--num_records=10 \
--scale=10 \
--image_type channels
The output of show_examples rasterizes each encoded variant example, and clearly shows the new we added (Figure 3). It worked!
Training with a Custom Channel
Now that we have determined that our new channel works, we can use it to train a new model. We will use data from the WES case study, but closely follow the detailed training instructions of the advanced training case study, highlighting here the most important aspects that pertain to the inclusion of our channel.
TRUTH_VCF="/benchmark/HG003_GRCh38_1_22_v4.2.1_benchmark.vcf.gz"
TRUTH_BED="/benchmark/HG003_GRCh38_1_22_v4.2.1_benchmark_noinconsistent.bed"
First, we run make_examples again in training mode, which will properly label the examples based on the truth set and confident regions we provide. This allows us to generate a training and validation set, using different chromosomal regions.
docker run \
-v "${PWD}/input":"/input" \
-v "${PWD}/output":"/output" \
-v "${PWD}/reference":"/reference" \
-v "${PWD}/benchmark":"/benchmark" \
${DOCKER_IMAGE} \
/opt/deepvariant/bin/make_examples \
--mode training \
--ref ${REF} \
--reads ${BAM} \
--truth_variants "${TRUTH_VCF}" \
--confident_regions "${TRUTH_BED}" \
--examples "output//training_set.with_label.tfrecord@.gz" \
--channels ${CHANNEL} \
--regions "'chr1'" \
--logtostderr
Following the same scheme of shuffling our examples, we produce a dataset_config.pbtxt file for each training and validation set.
TRAINING_DIR=/output/training_dir
DATASET_CONFIG_PBTXT="/output/training_set.dataset_config.pbtxt"
mkdir -p ${PWD}${TRAINING_DIR}
Now model_train can use this training data with our new custom channel and train its deep neural net.
docker run \
-v "${PWD}/output":"/output" \
${DOCKER_IMAGE} \
/opt/deepvariant/bin/model_train \
--dataset_config_pbtxt="${DATASET_CONFIG_PBTXT}" \
--train_dir="${TRAINING_DIR}" \
--model_name="${CHANNEL}_model" \
--number_of_steps=50000 \
--save_interval_secs=300 \
--batch_size=32 \
--learning_rate=0.0005
It is possible to set the flags –start_from_checkpoint and –allow_warmstart_from_different_num_channels to start from an existing model checkpoint trained on a different channel configuration. This may significantly reduce the amount of additional training required.
As outlined in our training guide, we can run model_eval simultaneously to determine the best model checkpoint. Once the training is done, we make sure to select our best performing model checkpoint.
MODEL_CHECKPOINT_PATH=$(cat "${TRAINING_DIR}"/best_checkpoint.txt)
Running a Model with a New Custom Channel
With our newly trained model ready, we can now run DeepVariant to perform variant calling. Since we excluded chromosome 20 in our training set and validation sets, we can use it here to test the model’s accuracy.
docker run \
-v "${PWD}/output":"/output" \
${DOCKER_IMAGE} \
/opt/deepvariant/bin/run_deepvariant \
--model_type WES \
--customized_model "${MODEL_CHECKPOINT_PATH}" \
--make_examples_extra_args=channels="${CHANNEL}" \
--ref "${REF}" \
--reads "${BAM}" \
--regions "chr20" \
--output_vcf "/output/test_set.vcf.gz"
Finally we may run the accuracy analysis as outlined in the WES case study using hap.py. If the data encoded in our new channel provides pertinent sequence information, we may reasonably expect some improvement in DeepVariant’s ability to correctly call variants.
Conclusion
Custom channels are a powerful way to further improve DeepVariant’s variant calling performance and tailor models for specific sequencing platforms or applications. We hope you will be able to deploy custom channels to further improve DeepVariant performance.

